Plasmid DNA is a versatile tool in molecular biology, often used for various genetic manipulations and studies. Restriction enzyme digestion of plasmid DNA is a crucial technique that allows researchers to precisely cleave DNA at specific sites, facilitating downstream applications such as cloning, sequencing and mapping. In this article, we will delve into the principles, procedures, reagents, and tips associated with restriction enzyme digestion of plasmid DNA.
Principle of digestion with restriction enzymes
Restriction digestion, also known as rrestriction endonuclease digestion, involves the cleavage of DNA at specific recognition sites using restriction enzymes. These enzymes recognize short palindromic sequences of nucleotides and catalyze the hydrolysis of phosphodiester bonds in the DNA backbone. This results in the formation of fragments with blunt or sticky ends, depending on the enzyme and its cutting mechanism.
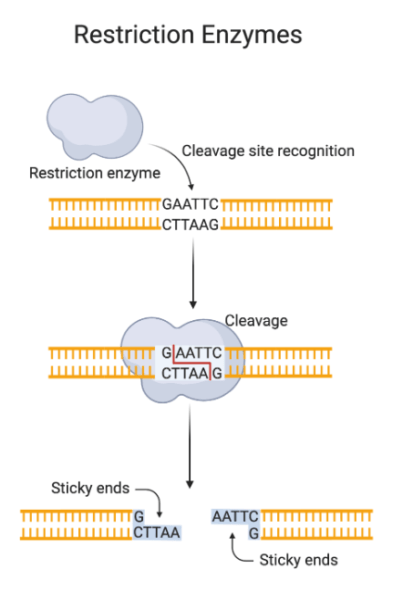
Importance and applications
Restriction digestion of plasmid DNA is essential for various molecular biology techniques, including:
- Molecular cloning: Restriction digestion allows the preparation of DNA fragments that can subsequently be ligated into vectors for cloning purposes.
- Sequence analysis: It provides a method to indirectly obtain sequence information by analyzing fragment patterns generated after digestion.
- Diagnostic Summaries: It is used to quickly check the identity of a plasmid by checking for the presence or absence of specific restriction sites.
Necessary reagents
To carry out restriction enzyme digestion of plasmid DNA, the following reagents are normally used:
- Plasmid DNA: The DNA template to be cleaved.
- Restriction enzymes): Enzymes that recognize and cleave specific DNA sequences.
- Constraint buffer: Provides optimal pH and ionic conditions for enzyme activity.
- BSA (bovine serum albumin): Optional, used to stabilize certain restriction enzymes.
- Gel Loading Dye and Electrophoresis Buffer: To analyze the digested DNA fragments by gel electrophoresis.
Plasmid DNA restriction enzyme digestion procedure
1. Selection of Restriction Enzymes:
Choose appropriate restriction enzymes based on the desired cleavage sites and the nature of the DNA fragment required.
Pro tip: Use sequence analysis tools to accurately predict enzyme cleavage sites.
2. Preparation of the reaction mixture:
Combine plasmid DNA, restriction enzymes, buffer, and any additional components such as BSA in a microcentrifuge tube.
Pro tip: Determine the appropriate reaction buffer and enzyme concentration according to the manufacturer's instructions.
3. Incubation:
Incubate the reaction mixture at the optimal temperature for enzymatic activity, usually 37 °C, for a specific period.
Pro tip: Ensure adequate incubation time for complete digestion, considering factors such as DNA concentration and enzyme activity.
4. Analysis:
After incubation, analyze the digested DNA fragments by gel electrophoresis to visualize the cleavage pattern and fragment sizes.
Tips for Successful Digestion of Plasmid DNA with Restriction Enzymes
- Proper management of enzymes: Ensure enzymes are stored and handled according to manufacturer's instructions to maintain activity.
- Optimization of reaction conditions: Adjust buffer conditions, enzyme concentration, and incubation time to achieve optimal digestion efficiency.
- Consideration of sensitivity to methylation: Choose enzymes that are compatible with the methylation status of the DNA.
- QA: Include positive and negative controls in each experiment to validate digestion efficiency.
- Documentation and analysis: Document experimental details and analyze gel images accurately to effectively interpret results.
Conclusion
Restriction enzyme digestion of plasmid DNA is a fundamental technique in molecular biology, allowing precise manipulation and analysis of genetic material. By understanding the principles, optimizing experimental conditions, and following best practices, researchers can successfully employ this technique for a wide range of applications, advancing our understanding of genetics and biotechnology. In summary, restriction digestion of plasmid DNA serves as a cornerstone in molecular biology research, allowing scientists to explore and manipulate the complexities of the genetic code. Through careful experimentation and innovation, this technique continues to drive advances in biotechnology and beyond.
Troubleshooting Plasmid DNA Digestion
Plasmid DNA digestion, a critical step in molecular biology experiments, can sometimes encounter challenges leading to incomplete or failed cleavage. Understanding potential problems and implementing effective problem-solving strategies is essential for reliable results. In this guide, we will explore common problems encountered during plasmid DNA digestion and provide practical solutions to address them.
Problem: incomplete digestion
Possible causes:
- Suboptimal incubation conditions: Incorrect temperature or duration of incubation.
- Enzymatic inactivation: Denaturation of the enzyme due to improper handling or storage.
- Suboptimal buffer conditions: buffer pH, salt concentration, or presence of incorrect inhibitors.
Solutions:
- Optimize incubation conditions: Ensure the reaction is maintained at the optimal temperature for the specified period according to the manufacturer's instructions.
- Handle enzymes with care: Store enzymes properly at recommended temperatures and avoid excessive freeze-thaw cycles. Use an ice bucket immediately after removing it from the freezer to prevent denaturation.
- Check buffer compatibility: Make sure the buffer used is suitable for your chosen enzymes and adjust the pH or salt concentration if necessary.
Problem: stellar activity
Possible causes:
- High concentration of glycerol: The presence of glycerol in the reaction buffer can induce non-specific cleavage.
- Suboptimal reaction conditions: Enzymatic activity influenced by variations in temperature, pH or ionic strength.
Solutions:
- Optimize reaction conditions: Maintain consistent reaction conditions, including temperature and buffer composition, to minimize non-specific cleavage.
- Use new tampons: Prepare fresh reaction buffers without excess glycerol to avoid unwanted enzymatic activity.
Problem: sensitivity to methylation
Possible causes:
- DNA methylation: The presence of methyl groups on specific DNA bases can inhibit enzymatic cleavage.
- Enzyme methylation: Some restriction enzymes can be sensitive to methylation, affecting their activity.
Solutions:
- Use methylation-sensitive enzymes: Select enzymes that are compatible with the methylation status of the DNA sample.
- Prevent methylation: Perform DNA isolation from bacterial strains that lack methylase activity or use specific enzymes sensitive to methylation.
Problem: contaminants or inhibitors
Possible causes:
- Contaminants in the DNA sample: The presence of contaminants such as phenol, chloroform or salts can interfere with enzymatic activity.
- Presence of detergents: Detergents in DNA isolation kits can inhibit enzymatic activity.
Solutions:
- Purify DNA sample: Use purification methods to remove contaminants before digestion.
- Avoid detergents: If you use commercial DNA isolation kits, be sure to wash them thoroughly to remove residual detergents.
Problem: poor ligation efficiency
Possible causes:
- Blunt Tip Fragments: DNA fragments with blunt ends may have lower ligation efficiency compared to fragments with cohesive ends.
- Mismatched adhesive ends: Incompatible adhesive ends can lead to inefficient ligation.
Solutions:
- Consider Cantilever Compatibility: Choose enzymes that generate compatible sticky ends for efficient ligation.
- Use T4 DNA ligase: Use T4 DNA ligase, which can ligate blunt and cohesive ends, to improve ligation efficiency.
Conclusion
Troubleshooting plasmid DNA digestion requires a systematic approach to identifying and addressing potential problems. By understanding the underlying causes of incomplete digestion or inefficient ligation, researchers can implement appropriate solutions to optimize experimental results. Through careful optimization of reaction conditions, enzyme selection, and sample preparation techniques, reliable digestion of plasmid DNA can be achieved, confidently facilitating subsequent molecular biology applications.
References
- New England Biolabs (NEB). (North Dakota). Digestions with restriction enzymes: protocol. Retrieved from NEB website
- Addgene. (North Dakota). Sequence analyzer. Retrieved from Addgene website
- Roberts RJ. (2005). How restriction enzymes became the workhorses of molecular biology. Proceedings of the National Academy of Sciences of the United States of America, 102(17), 5905–5908. https://doi.org/10.1073/pnas.0500923102
- Wilson GG, Murray NE. (1991). Restriction and modification systems. Annual Review of Genetics, 25, 585–627.
- Pingoud A, Jeltsch A. (2001). Structure and function of type II restriction endonucleases. Nucleic Acid Research, 29(18), 3705–3727.
- Sambrook J, Russell DW. (2001). Molecular cloning: laboratory manual (3rd ed.). Cold Spring Harbor Laboratory Press.













Leave feedback about this